This vignette documents the steps to ingest model output files (in CSV and NetCDF format) for use in the dashboard.
This ETL was validated against two analyses received from IWMI scientists, one for Mali (Niger river basin), and another for Kenya (Mara river basin). The water accounting models are written in Python and the code repositories are on GitHub (e.g. WAPORWA notebooks for the Mali analysis).
The results of the WA+ balances consist in time-series of constructed hydrological variables – the time steps can vary (dekadal, monthly, seasonal, yearly steps). The series are either gridded (NetCDF) or aggregated over an entire zone of interest (CSV output files). The modeled variables are further enriched to construct visual balances (or “Sheets”) that represent key water availability, use and management indicators over time.
The WA+ modeling approach can vary slightly across use cases (different temporal extent, number of sub-basins, different flow units, different mix of input layers and output variables). To the extent possible this ETL intends to produce a generalized data model.
The steps outlined below serve as a blueprint for a generic data_etl() method included in this package (refer to the method’s documentation for more details, esp. how to ingest additional result sets). The objective is to generate a data cube suitable for online analytics.
# Location of model output datasets
dir <- getOption("wa.data")
# Plot defaults
th <- function(...) theme(
legend.position = "none",
panel.spacing.y = unit(0, "lines"),
strip.text.y = element_text(size=7, angle=0, hjust=0),
strip.background = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank(),
axis.ticks.y = element_blank(),
panel.border = element_blank(),
panel.grid = element_blank(),
panel.background = element_blank(),
...)Basin-level Aggregates (CSV files)
We start below by inventorying all basin-level variables contained in the modeled CSV files.
# List all output CSV
data <- list(
ken = file.path(dir, "./ken/hydroloop_results/csv"),
mli = file.path(dir, "/mli/csv_km3")
) %>%
lapply(list.files, pattern="*.csv", recursive=TRUE, full.names=TRUE) %>%
lapply(data.table) %>%
rbindlist(idcol="iso3", use.names=TRUE, fill=TRUE) %>%
setnames("V1", "path")
# Extract timestamps and sheet codes from file names
data[, `:=`(
file = basename(path)
)][, `:=`(
year = str_extract(file, "_[0-9]{4}") %>% str_sub(2,5) %>% as.integer(),
month = str_extract(file, "[0-9]{4}_[0-9]{1,2}") %>% str_sub(6,7) %>% as.integer(),
sheet = str_extract(tolower(file), "sheet[0-9]{1}")
)] %>% setorder(iso3, sheet, year, month, na.last=TRUE)
# Append flow units (typically km3 or MCM, will need to be specified manually)
data[, unit := fcase(
iso3=="mli", "km3",
iso3=="ken", "km3"
)]We have 8 yearly water balances for Mali (Sheet #1 variables), 15 yearly balances for Kenya (across 6 sheets), as well as monthly and seasonal breakdowns.
data[iso3=="mli", .(iso3, file, sheet, year, month)] %>%
paged_table()
data[iso3=="ken" & !is.na(year) & is.na(month), .(iso3, file, sheet, year, month)] %>%
paged_table()
data[iso3=="ken" & !is.na(month), .(iso3, file, sheet, year, month)] %>%
paged_table()The Kenya analysis also includes yearly, seasonal, and monthly time-series for an additional 57 variables:
Variable Inspection
We also need to validate naming conventions and units for all output variables, so we continue by inspecting individual files starting with Mali.
Sheet #1
f <- data[iso3=="mli"]
mli <- lapply(1:nrow(f), function(x) fread(f[x, path])[, `:=`(
iso3 = f[x, iso3],
sheet = f[x, sheet],
year = f[x, year]
)]) %>% rbindlist()
setnames(mli, tolower(names(mli)))
mli %>% paged_table(options=list(max.print=300))Output for Sheet #1 include 34 unique variables categorized into class and subclass. The naming is consistent across years.
mli %>%
ggplot() +
geom_line(aes(year, value, color=paste(class, subclass))) +
facet_grid(paste(class, subclass, variable)~., scales="free_y") +
th() +
labs(x=NULL, y=NULL) +
ggtitle("WA+ Sheet 1 Variables - Mali, 2009 to 2016")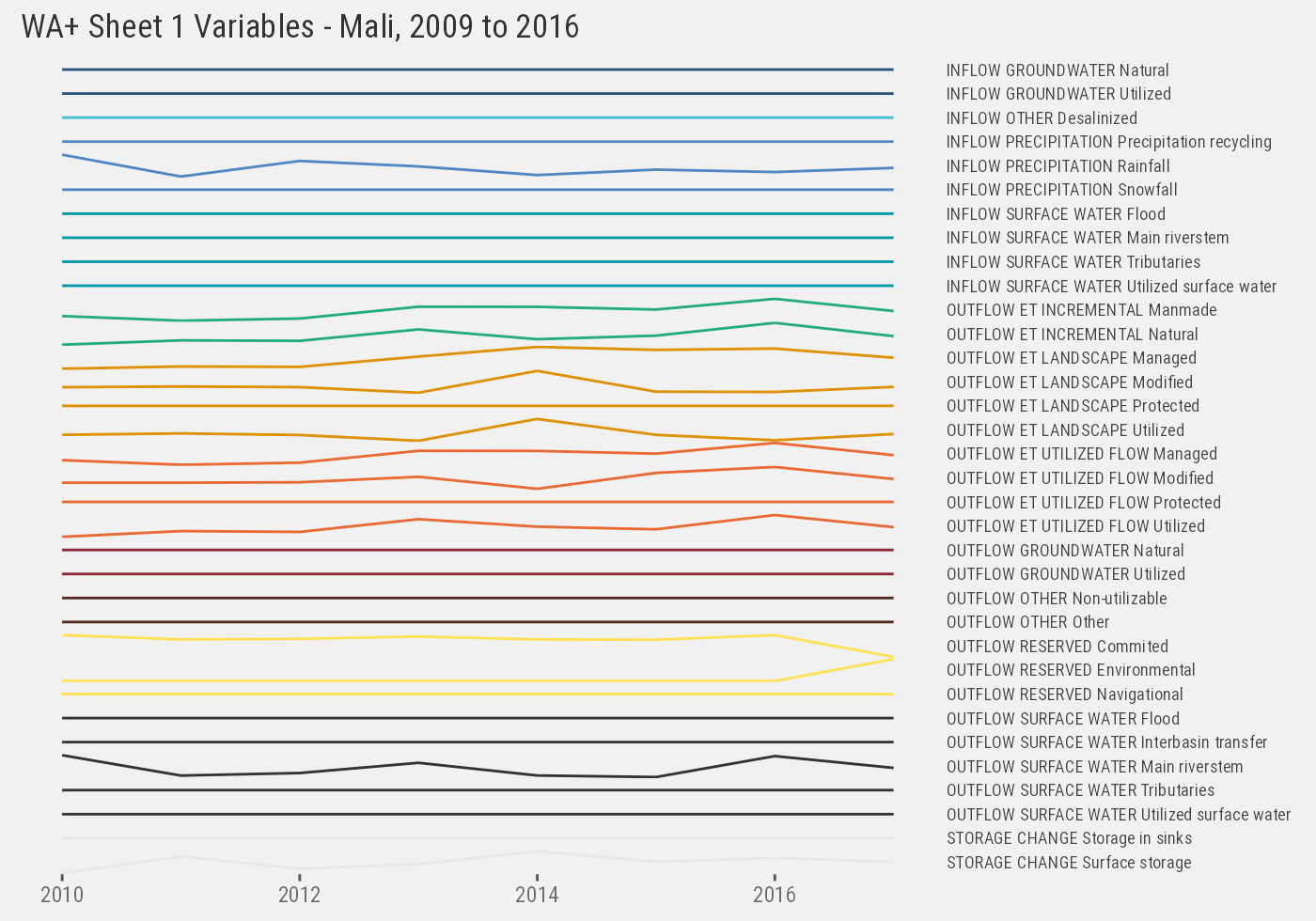
Similarly we look at Kenya’s monthly time-series. First at Sheet #1 variables:
f <- data[iso3=="ken" & !is.na(month) & !is.na(year) & sheet=="sheet1"]
ken_1 <- lapply(1:nrow(f), function(x) fread(f[x, path])[, `:=`(
iso3 = f[x, iso3],
sheet = f[x, sheet],
year = f[x, year],
month = f[x, month]
)]) %>% rbindlist()
setnames(ken_1, tolower(names(ken_1)))
ken_1 %>% paged_table(options=list(max.print=300))
ken_1 %>%
ggplot() +
geom_line(aes(as.Date(paste(year, month, 1), "%Y %m %d"), value, color=subclass)) +
facet_grid(paste(class, subclass, variable)~., scales="free_y") +
th() +
labs(x=NULL, y=NULL) +
ggtitle("WA+ Sheet 1 Variables - Kenya, 2003 to 2017")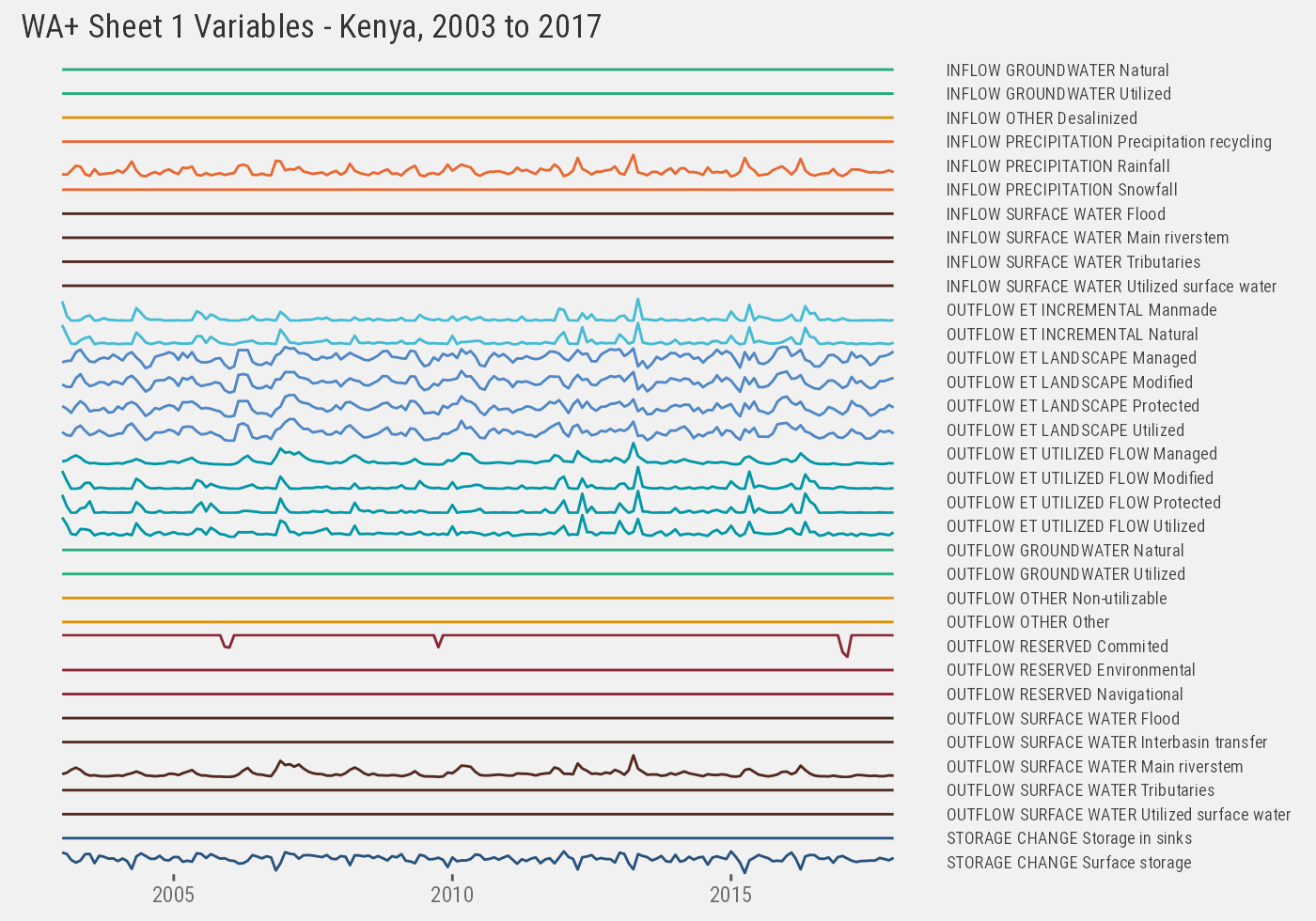
Sheet #2
Then we extract Sheet #2 variables (this is a breakdown of evapotranspiration by land use classes):
f <- data[iso3=="ken" & !is.na(month) & !is.na(year) & sheet=="sheet2"]
ken_2 <- lapply(1:nrow(f), function(x) fread(f[x, path])[, `:=`(
iso3 = f[x, iso3],
sheet = f[x, sheet],
year = f[x, year],
month = f[x, month]
)]) %>% rbindlist()
setnames(ken_2, tolower(names(ken_2)))
ken_2 %>% paged_table(options=list(max.print=300))Each sheet uses a different data structure, but they can be normalized into a “long” format:
# Normalize
ken_2 <- melt(ken_2, id.vars=c("land_use", "class", "iso3", "sheet", "year", "month"),
variable.factor=FALSE)
# Standardize category names
setnames(ken_2, c("land_use", "class"), c("class", "subclass"))
setorder(ken_2, class, subclass, variable, year, month)We only plot variables for the first land use class for illustration purposes:
ken_2[class %in% ken_2[, unique(class)][1]] %>%
ggplot() +
geom_line(aes(as.Date(paste(year, month, 1), "%Y %m %d"), value, color=subclass)) +
facet_grid(paste(class, subclass, variable)~., scales="free_y") +
th() +
labs(x=NULL, y=NULL) +
ggtitle("WA+ Sheet 2 Variables - Kenya, 2003 to 2017")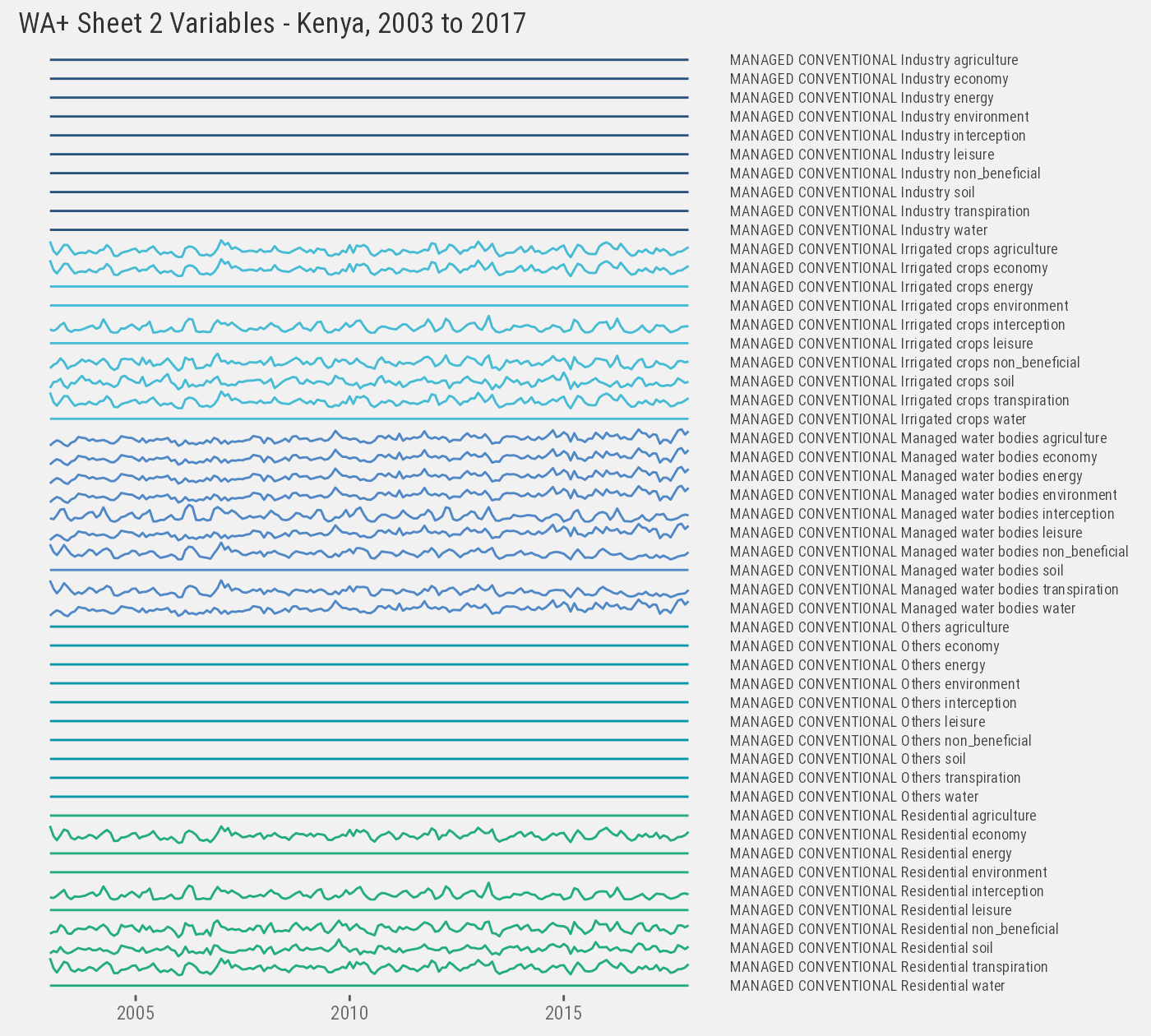
Sheet #3
Similarly we inspect results for Sheet #3 “Agriculture Services”. In the Kenya analysis, variables for Sheet #3 to Sheet #6 are available as yearly, seasonal, and/or monthly time-series (across multiple CSV files).
We (arbitrarily) gather all the seasonal time-series1:
f <- data[iso3=="ken" & sheet=="sheet3" &
str_detect(file, "season") & str_detect(file, "54.0")]
ken_3 <- lapply(1:nrow(f), function(x) fread(f[x, path])[, `:=`(
iso3 = f[x, iso3],
sheet = f[x, sheet],
year = f[x, year],
month = f[x, month],
variable = str_replace(f[x, file], "sheet3_54.0_", "") %>% str_replace("_season.csv", "")
)]) %>% rbindlist()
ken_3[, `:=`(
# Drop counter column
V1 = NULL,
# Ensure end of season is last day of month (for consistency with other sheets)
end_dates = end_dates - 1
)][, `:=`(
# Encode year/month columns using end of season
year = year(end_dates),
month = month(end_dates)
)]
setnames(ken_3, "Seasonal", "value")
setnames(ken_3, c("start_dates", "end_dates"), c("date_start", "date_end"))
setorder(ken_3, variable, year, month)
ken_3 %>% paged_table(options=list(max.print=300))Plotting the entire series (6 variables):
ken_3 %>%
ggplot() +
geom_line(aes(as.Date(paste(year, month, 1), "%Y %m %d"), value, color=variable)) +
facet_grid(variable~., scales="free_y") +
th() +
labs(x=NULL, y=NULL) +
ggtitle("WA+ Sheet 3 Variables - Kenya, 2003 to 2017 (seasonal)")## Warning: Removed 24 row(s) containing missing values (geom_path).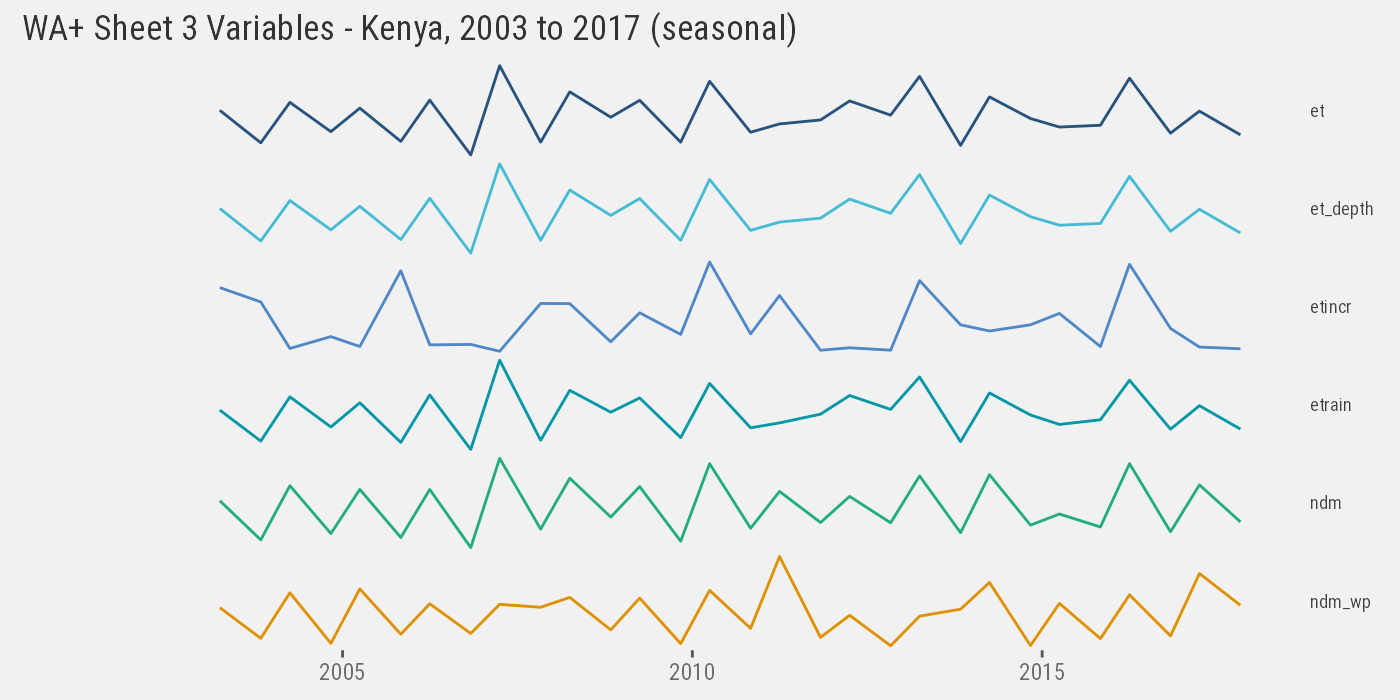
We gather all Sheet #1 to Sheet #3 variables above into a long table format.
cube <- rbind(mli, ken_1, ken_2, ken_3, fill=TRUE)
setcolorder(cube,
c("iso3", "sheet", "class", "subclass", "variable",
"year", "month", "date_start", "date_end"))For consistency’s sake, we encode date_start and date_end in all sheets (incl. yearly and monthly variables):
cube[is.na(date_start) & is.na(month), `:=`(
# yearly series: 1/1 to 12/31
date_start = as.IDate(paste(year, 1, 1), "%Y %m %d"),
date_end = as.IDate(paste(year, 12, 31), "%Y %m %d")
)][is.na(date_start) & !is.na(month), `:=`(
# monthly series: 1/1 to 1/31
date_start = as.IDate(paste(year, month, 1), "%Y %m %d"),
date_end = ceiling_date(as.IDate(paste(year, month, 20), "%Y %m %d"), "months") - days(1)
)]
#saveRDS(cube, "./data-raw/rds/data.rds")The generated data cube includes the following dimensions:
- iso3
- sheet
- class
- subclass
- variable
- year
- month
- date_start
- date_end
- value
Data Dictionaries
Another ETL step is required to map the variable codes used in the data files to their visual representation in the WA+ Sheets (SVG designs).
knitr::include_graphics(file.path("./fig", list.files("./vignettes/fig")))This mapping is embedded in the WA+ print_sheet Python module, (example for Sheet 1 in the WAPORWA repo). Importantly many SVG elements are fields calculated from output variables. We might need to replicate that logic in the dashboard to create dynamic visuals2.
Sheet #1
Field mappings are constructed in the table below.
s1 <- fread(system.file("./csv/sheet_1_schema.csv", package="WADashboard")) %>%
paged_table()Formulas for calculated fields are replicated here.
We proceed similarly for Sheet #2 and Sheet #3 mappings.